7 Metadata - Darwin Core
The Darwin Core (DwC) is a body of standards maintained by TDWG (Biodiversity Information Standards, formerly known as The International Working Group on Taxonomic Databases). It includes a glossary of terms intended to facilitate the sharing of information about biological diversity by providing reference definitions, examples, and commentaries. The Darwin Core is primarily based on taxa, their occurrence in nature as documented by observations, specimens, and samples, and related information.
Darwin Core revolves around a standard file format, the Darwin Core Archive (DwC-A). This compact package (a ZIP file) contains interconnected text files and enables data publishers to share their data using a common terminology. DwC is used both by GBIF and OBIS for publishing data.
This standardization not only simplifies the process of publishing biodiversity datasets, it also makes it easy for users to discover, search, evaluate and compare datasets as they seek answers to today’s data-intensive research and policy questions.
7.1 Darwin Core structure
A DWC-A can contains 4 types of components. These files can include:
- eml.xml
- core file
- extension files (optional)
- meta.xml
EML stands for Ecological Metadata Language. This file contains the dataset metadata in an XML format.
Core and extension files contain data records, arranged one per line. Each row in the extension file or extension record points to a single record in the core file. For each single core record there can be many extension records. This is sometimes referred to as a “star schema”.
The meta.xml file describes how the files in the archive are organised. It describes the linkage between the core and extension files and maps non-standards column names to Darwin Core terms.
To publish the data as a DwC-A, we recommend to upload the core and extension files to the IPT (e.g. IPT.biodiversity.aq). Use the IPT’s built-in metadata editor to enter dataset metadata. The IPT will compile the EML on the data you provided, as well as the meta.xml.
7.2 Types of Darwin Core archives
- Resource Metadata - Used to describe a biodiversity information resource, including contact details, even if no digital data can currently be shared (this provides a way for researchers to discover resources which are not yet available online).
- Checklist - Used to share annotated species checklists, taxonomic catalogues, and other information about taxa.
- Occurrence - Used to share information about a specific instance of a taxon such as a specimen or observation.
- Event - Used to share information about the protocols used in ecological investigations.
7.3 Darwin Core details
In general, metadata EML will accompany some data core in a DwC-A. It provides a description and details on the resource.
It is also possible to initially only publish your metadata and add your data at a later stage.
This allows researchers to discover resources that or not yet available online. It is best to consider your abstract as the material and methods section of your paper but then for just your data. Try to provide as much detail as possible.
Actually if you fill out your metadata extensively you have a first very good draft for a data paper (if that is something you would like to make). You’ll just have to add some maps, summary statistics, and you are good to go.
If you are in a hurry, below is an overview of what currently are the required fields.
If you use the IPT to create your Darwin Core archive, you will have to choose a short name for your dataset. Choose this wisely because it can’t be changed. The short name serves as an identifier within the IPT installation, and will be used as a parameter in the URL to access the resource via the Internet. For the short name you can only use alphanumeric characters, hyphens, or underscores.
7.3.1 Required metadata fields:
7.3.1.1 Basic Metadata
7.3.1.1.1 title
This will be the long title of your dataset, and how the dataset will be cited
7.3.1.1.2 publishing organisation
Please select the organisation responsible for publishing (producing, releasing, holding) this resource. It will be used as the resource’s publishing organisation when registering this resource with GBIF and when submitting metadata during DOI registrations. It will also be used to auto-generate the citation for the resource (if auto-generation is turned on), so consider the prominence of the role. Please be aware your selection cannot be changed after the resource has been either registered with GBIF or assigned a DOI.
On our IPT you can choose
- Antarctic Biodiversity Information Facility (AntaBIF) for terrestrial datasets
- SCAR - AntOBIS for marine datasets
- SCAR - Microbial Antarctic Resource System for microbial datasets
We can also publish on behalf of others. Currently we have agreements with
- British Antarctic Survey
- Italian Antarctic National Museum (MNA, section Genua)
7.3.1.1.3 type
The type of resource. The value of this field depends on the core mapping of the resource and is no longer editable if the Darwin Core mapping has already been made.
This can be
- Metadata-only
- Checklist
- Occurence
- Event-Core
7.3.1.1.4 metadata language
The language in which the metadata document is written.
7.3.1.1.5 data language
The primary language in which the described data (not the metadata document) is written.
7.3.1.1.6 Update Frequency
The frequency with which changes are made to the resource after the initial resource has been published. For convenience, its value will default to the auto-publishing interval (if auto-publishing has been turned on), however, it can always be overridden later. Please note a description of the maintenance frequency of the resource can also be entered on the Additional Metadata page.
7.3.1.1.7 data license
The licence that you apply to a dataset provides a standardized way to define appropriate uses of your work. GBIF encourages publishers to adopt the least restrictive licence possible from among three machine-readable options (CC0 1.0, CC-BY 4.0 or CC-BY-NC 4.0) to encourage the widest possible use and application of data. Learn more here. If you are unable to choose one of the three options, and your dataset contains occurrence data, you will not be able to register your dataset with GBIF or make it globally discoverable through GBIF.org. If you feel unable to select one of the three options, please contact the GBIF Secretariat at participation@gbif.org.
7.3.1.1.8 description
A brief overview of the resource that is being documented broken into paragraphs.
Think of it as an abstract for a (data) paper
7.3.1.1.9 Resource contact(s)
7.3.1.1.10 Resource creator(s)
7.3.1.1.11 metadata provider(s)
- Last name
- Position
- Organisation
7.3.1.2 Geographic coverage
7.3.1.2.1 description
7.3.2 Citation auto generation
Creator 1 R, Creator 2 R, Creator 3 R (2019): How to create a metadata record. v1. Publishing organisation. Dataset/Type. https://ipt.biodiversity.aq/resource?r=shortname&v=1.0
7.3.2.1 Example
Griffiths H J, Linse K, Crame J (2017): SOMBASE – Southern Ocean mollusc database: a tool for biogeographic analysis in diversity and evolution. v1.6. British Antarctic Survey. Dataset/Occurrence. https://ipt.biodiversity.aq/resource?r=sombase_southern_ocean_mollusc_database&v=1.6
7.4 Metadata for occurrence data
In general, the more information you provide the more knowledge can be gained from your occurrence data.
The first user of this data is future yourself. Taking the time to be as detailed in providing and describing your data is time you will gain in the future when you want to reanalyse your data or integrate it in a broader study.
The best way is to save the data in some tabular format, something like a csv-file or tab-delimted txt. It is not recomended to use Excel, because it has the habit of giving you unwanted help, resulting in messed up data…
Here we start with a description of the minimal metadata that you should provide in order to analyse your data, including developing a species distribution modelling.
Each row in your dataset should corresponding to an observation. The names of the columns in the dataset should correspond to DwC terms.
You can find a full description of the Darwin Core terms in the Darwin Core quick reference guide. But below we provide an overview of the most important terms. If you have information that doesn’t ft the terms below just check the Darwin Core quick reference guide or create an issue on the EG-ABI course github or the TDWG Github.
Your metadata should allow you or anybody else to determine What, Where, When, Who and How.
The Darwin Core Standards has 8 absolutely required terms. basisOfRecord, occurrenceID, scientificName, scientificNameID, occurrenceStatus, decimalLongitude, decimalLatitude, eventDate
Besides those we recommend a number of other ones as well. In general it is better to provide as many as you can.
basisOfRecord
- type
- associatedReferences
occurrenceID
- institutionCode
- collectionCode
- catalogNumber
scientificName, scientificNameID
- scientificNameAuthorship
- IdentificationQualifier
- taxonrank
occurrenceStatus
- organismQuantity
- organismQuantityType
- sex
- lifeStage
- behavior
- occurrenceRemarks
- identifiedBy
- dateIdentified
decimalLongitude, decimalLatitude
- geodeticDatum
- coordinateUncertaintyInMeters
eventDate
- year
- month
- Day
- Time
- Timezone
- samplingProtocol
7.4.1 What
7.4.1.1 basisOfRecord
Additional term
basisOfRecord describes the nature of record. It has a to follow a specific vocabulary. Some of these might overlap sometimes. That is why it is useful to use type as well.
- PreservedSpecimen A specimen that has been preserved. This can be a museum specimen, a plant on an herbarium sheet, fish in a bag in a freezer.
- FossilSpecimen A resource describing a fossilized specimen.
- LivingSpecimen A resource describing a living specimen. For instance an animal in a zoo.
- MaterialSample A resource describing a material sample. For instance a material sample from an organism in the wild and sample put into a collection (blood sample, biopsy).
- HumanObservation A resource describing an observation made by one or more people. For instance an organism observed in the wild and written in field notes. Samples that were collected during an expedition but thrown back into the sea.
- MachineObservation An output of a machine observation process. Any kind of automated sensor like camera traps.
- Occurrence A resource describing an instance of the Occurrence class. NOTE: this value is ambiguous and hence should only be used when the resource type is unknown.
The nature or genre of the resource. Must be populated with a value from the DCMI type vocabulary.
Options include: Image, StillImage, MovingImage, Interactive Resource, PhysicalObject, Sound, Text, PhysicalObject, Collection, Dataset, Event, Service, Software.
associatedReferences A list (concatenated and separated) of identifiers (publication, bibliographic reference, global unique identifier, URI) of literature associated with the occurrence.
Example: If you want to add a record from literature you should use basisOfRecord:HumanObservation with type:Text in combination with associatedReferences:Bibliographic reference
7.4.1.2 occurrenceID
Additional Terms
institutionCode, collectionCode, catalogNumber
occurrenceID is an identifier for the occurrence record and should be globally unique and persistent. In the absence of a persistent global unique identifier, construct one from a combination of identifiers in the record that will most closely make the occurrenceID globally unique. Something like the combination of institutionCode, collectionCode, and catalogNumber is a generally used format.
There are a number of other terms that you could use as well like datasetName and datasetID.
institutionCode The name (or acronym) in use by the institution having custody of the object(s) or information referred to in the record.
institutionID An identifier for the institution having custody of the object(s) or information referred to in the record.
For physical specimens, the recommended best practice is to use a code and identifier from a collections registry such as the Global Registry of Biodiversity Repositories (http://grbio.org/).
| Institution code | Institution name |
|---|---|
| AAS | British Antarctic Survey |
| BRM | Alfred-Wegener-Institut für Polar- und Meeresforschung |
| MNA | The Museo Nazionale dell’Antartide (Italian National Antarctic Museum in Genoa) |
| RBINS | Royal Belgian Institute of Natural Sciences |
collectionCode The name, acronym, coden, or initialism identifying the collection or data set from which the record was derived.
datasetName The name identifying the data set from which the record was derived.
datasetID An identifier for the set of data. May be a globally unique identifier or an identifier specific to a collection or institution.
catalogNumber An identifier (preferably unique) for the record within the data set or collection.
recordNumber An identifier given to the occurrence at the time it was recorded. Often serves as a link between field notes and an occurrence record, such as a specimen collector’s number.
| occurrenceID | institutionCode | collectionCode | catalogNumber |
|---|---|---|---|
| http://arctos.database.museum/guid/MSB:Mamm:233627 | MSB | Mamm | 233627 |
| PS89_FF_000023 | PS89 | FF | 000023 |
It is a good practice to keep your catalogue/record numbers of equal length throughout your dataset.
To keep it persistent just don’t change it. If you reference another source, keep the number.
For example if you have samples that were collected during a marine expedition, usually all sampling events get ther own ID. Don’t invent a new ID for the event. This will help you to find any information related to that event at a later stage.
7.4.1.3 scientificName and scientificNameID
Additional Terms
Kingdom, taxonrank, scientificNameAuthorship, IdentificationQualifier
Additional Additional Terms (See Who)
identifiedBy, dateIdentified, typeStatus
scientificName In the spirit of keeping things persistent this should always contain the originally recorded scientific name, even if the name is currently a synonym (if the sample has been reidentified this is of course a different case). The scientific name should always be to the lowest possible taxonomic rank of which you are certain. In case of uncertain identifications, and the scientific name contains qualifiers such as cf., ? or aff., then this name should go in identificationQualifier.
scientificNameAuthorship OBIS recommends to not include authorship in scientificName, and only use scientificNameAuthorship for that purpose.
scientificNameID Following the OBIS guidelines a WoRMS LSID should be added in scientificNameID. LSIDs are persistent, location-independent, resource identifiers for uniquely naming biologically significant resources. More information on LSIDs can be found at http://www.lsid.info.
taxonrank, Determines the level of identification. This can be for kingdom, phylum, class, order, family, genus, subgenus, specificEpithet, infraspecificEpithet. It is good practice to always provide at least the kingdom. In case you can provide a WoRMS LSID the others are not really required. Otherwise it would be good to add them if possible.
identificationQualifier A brief phrase or a standard term (“cf.”, “aff.”) to express the determiner’s doubts about the identification.
The use of confer meaning compare and abbeviated to cf. in a scientific name means that the person using the name is saying the animal should be compared to a given species, as it might not be exactly the same species. It’s a way of applying a provisional name to a species and is most frequently used when new fish are discovered that look slightly different to the form normally encountered. It indicates that the fish might be a variant of the same species, but could also turn out to be something completely different.
Species affinis (commonly abbreviated to: sp. aff., aff., or affin.) indicates that available material or evidence suggests that the proposed species is related to, has an affinity to, but is not identical to, the species with the binomial name that follows.
Species abbreviated as sp. commonly occurs when authors are confident that some individuals belong to a particular genus but are not sure to which exact species they belong.
Authors may also use “spp.” (Species pluralis) as a short way of saying that something applies to many species within a genus, but not to all.
| scientificName | scientificNameAuthorship | IdentificationQualifier | taxonrank | |
|---|---|---|---|---|
| 1 | Electrona | Goode & Bean, 1896 | aff. risso | genus |
| 2 | Electrona antarctica | Günther, 1878 | species | |
| 3 | Electrona | Goode & Bean, 1896 | sp. | genus |
| 4 | Electrona | Goode & Bean, 1896 | spp. | genus |
What do these examples mean?
- Case 1: “I collected something that looks like Electrona risso but I’m not sure.”
- Case 2: “I’m 100% sure this is Electrona antarctica)
- Case 3: “I’m sure about the genus, but it could be any one of multiple species”
- Case 4: “I have this bag with fish; I’m sure about the genus but it could be a variety of species within the genus”
7.4.1.4 occurrenceStatus
Additional Terms
organismQuantity, organismQuantityType, sex, lifeStage, behavior, occurrenceRemarks
occurrenceStatus is a statement about the presence or absence of a taxon at a location. It is an important term, because it allows us to distinguish between presence and absence records. It is an OBIS required term and should be filled in with either “present” or “absent”.
organismQuantity, organismQuantityType These two always go together. organismQuantity is a number for the quantity of animals and organismQuantityType defines the type of quantification system used for the quantity of organisms.
| organismQuantity | organismQuantityType |
|---|---|
| 27 | individuals |
| 12.5 | %biomass |
| r | BraunBlanquetScale |
There are a few other terms as well that we want to mention. The old term that was used was individualCount but that wasn’t really versatile. Still in some cases it can be useful to mention it.
Please take note that OBIS recommends all quantitative measurements and sampling facts to be treated in the ExtendedMeasurementOrFact extension and not in the Darwin Core files. OBIS recomends using ExtendedMeasurementOrFact in combination with Darwin Core Event Core, which is a little more advanced.
sexdecimalLatitude The sex of the biological individual(s) represented in the occurrence. For the OBIS-recommended vocabulary for sex see BODC vocab : S10
lifeStagedecimalLatitude The age class or life stage of the biological individual(s) at the time the occurrence was recorded. The OBIS recommended vocabulary for lifestage is BODC vocab: S11
behaviordecimalLatitude The behavior shown by the subject at the time the occurrence was recorded (no OBIS recomended vocab available).
occurrenceRemarksdecimalLatitude can hold any comments or notes about the occurrence.
7.4.2 Where
7.4.2.1 decimalLatitude anddecimalLongitude
Additional Terms
geodeticDatum, coordinateUncertaintyInMeters, locality, locationID, footprintWKT
To determine the location of a point on a globe you need at least the decimalLatitude and decimalLongitude and and a definition of the spatial reference system that was used. The spatial reference system defines the model of the earths shape that was used.
The system currently used by GPS is WGS84 (also known as WGS 1984, EPSG:4326). Quite often the geodeticDatum is not provided but it is actually essential.
decimalLatitude defines the position (in degrees) relative to the equator and ranges from 90 (North Pole) to -90 (South Pole).
decimalLongitude defines the position (in degrees) relative to the Greenwich Meridian and ranges from 180 to -180.
Mak sure not to switch those around. The standard notation has them as latitude followed by longitude. Some people naturally tend to write them as longitude followed by latitude, because this is conceptually similar to a cartesian (x-y) notation. However, especially for samples collected in the area of the Antarctic Peninsula, these switches can be hard to spot.
coordinateUncertaintyInMeters The horizontal distance (in meters) from the given decimalLatitude and decimalLongitude describing the smallest circle containing the whole of the location. Leave the value empty if the uncertainty is unknown, cannot be estimated, or is not applicable (because there are no coordinates). Zero is not a valid value for this term. This one is actually closely linked to the number of decimals in your latitude and longitude values.
In cases where decimal latitude are calculated they are often expressed with too many digits. If you have more than 6 in biology it doesn’t real make sense anymore or it doesn’t represent how precise the measurement was in the field.
To explain it, see this XKCD commic or the table below.
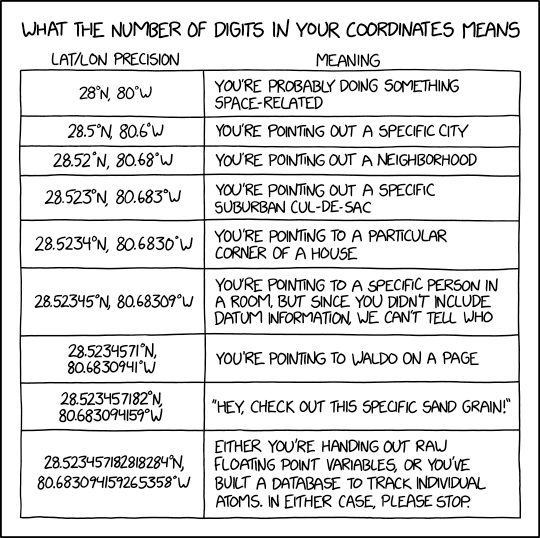
| decimal places | decimal degrees | DMS | Object that can be unambiguously recognized at this scale | N/S or E/W at equator | E/W at 67N/S |
|---|---|---|---|---|---|
| 0 | 1.0 | 1° 00’ 0" | country or large region | 111.32 km | 43.496 km |
| 1 | 0.1 | 0° 06’ 0" | large city or district | 11.132 km | 4.3496 km |
| 2 | 0.01 | 0° 00’ 36" | town or village | 1.1132 km | 434.96 m |
| 3 | 0.001 | 0° 00’ 3.6" | neighborhood, street | 111.32 m | 43.496 m |
| 4 | 0.0001 | 0° 00’ 0.36" | individual street, land parcel | 11.132 m | 4.3496 m |
| 5 | 0.00001 | 0° 00’ 0.036" | individual trees, door entrance | 1.1132 m | 434.96 mm |
| 6 | 0.000001 | 0° 00’ 0.0036" | individual humans | 111.32 mm | 43.496 mm |
| 7 | 0.0000001 | 0° 00’ 0.00036" | practical limit of commercial surveying | 11.132 mm | 4.3496 mm |
| 8 | 0.00000001 | 0° 00’ 0.000036" | specialized surveying (e.g. tectonic plate mapping) | 1.1132 mm | 434.96 µm |
7.4.3 When
7.4.3.1 eventDate
Additional Terms
year, month, Day, Time, Timezone
eventDate The date and time when the occurrence/event was recorded. Can be the date-time or interval during which an event occurred. This term uses the ISO 8601 standard. OBIS recommends using the extended ISO 8601 format with hyphens.

For time intervals ISO 8601 uses / as a separator. Date and time are separated by T. Times can have a time zone indicator at the end, if this is not the case then the time is assumed to be local time. When a time is UTC, a Z is added. Some examples of ISO 8601 dates are:
1973-02-28T15:25:00 2005-08-31T12:11+12 1993-01-26T04:39+12/1993-01-26T05:48+12 2008-04-25T09:53 1948-09-13 1993-01/02 1993-01 1993
It is an option to split out the eventDate into its seperate components. This can be a good safeguard against software like Excel reformatting your dates and times.
year, month, Day, Time, Timezone
verbatimEventDate You can put the original date format here, not really needed but might be useful, in case where you are digitising old literature records.
7.4.4 Who
A list (concatenated and separated) of names of people, groups, or organizations who assigned the Taxon to the subject. Recommended best practice is to separate the values in a list with space vertical bar space ( | )
This one is very seldom filled out but it is actually an important one and it should be specific. It should be the person that did the actual identification not the suprvisor.
In case of a reindentification it is often useful to be able to contact the person who did the initial identification.
7.4.5 How
This is a tricky one it is quite relevent but this field often doesn’t provide enough space to describe what you did. For describing a specific gear you can use BODC vocab : L22